March, 2024 | Frontiers in Plant Science |
Introduction: Developing climate-smart agriculture requires cost-effective methods to characterize crop growing conditions. A research team from the Americas, led by the University of São Paulo (Brazil), addresses a key bottleneck in climate-smart breeding: the lack of widely adopted envirotyping pipelines that can both guide field-trial design and improve genomic prediction accuracy. Using tropical maize multi-environment trials in Brazil, the authors introduce an enviromic assembly–aided genomic prediction (E-GP) framework grounded in Shelford’s Law of Tolerance. By translating raw environmental data into ecophysiologically meaningful typology markers, the study evaluates whether structured environmental information can improve the prediction of yield plasticity while substantially reducing the need for extensive in-field phenotyping.
Key findings: The study shows that E-GP consistently outperforms conventional genomic best linear unbiased prediction (GBLUP), particularly in predicting yield plasticity and performance in new or poorly characterized environments. In tropical maize trials, selective phenotyping guided by enviromic–genomic kinships achieved reliable predictions using only 1.5–4% of the original genotype–environment combinations. Incorporating enviromic assembly increased the accuracy of yield plasticity coefficients (b‚āĀ) by 437% (r = 0.43 versus r = 0.08 under GBLUP), while predictive ability for untested genotypes in nitrogen-limited environments was 118% higher than baseline models. Across scenarios, results demonstrate that the representativeness of genotype–environment combinations is more critical than the overall size of multi-environment trials (METs). By enabling accurate prediction with sharply reduced field phenotyping, enviromic assembly provides a parsimonious and biologically grounded platform for early screening of yield plasticity, offering regional breeding programs a cost-effective pathway to improve climate adaptability, particularly for resource-constrained breeding programs with limited capacity for extensive field trials or sensor-based envirotyping.
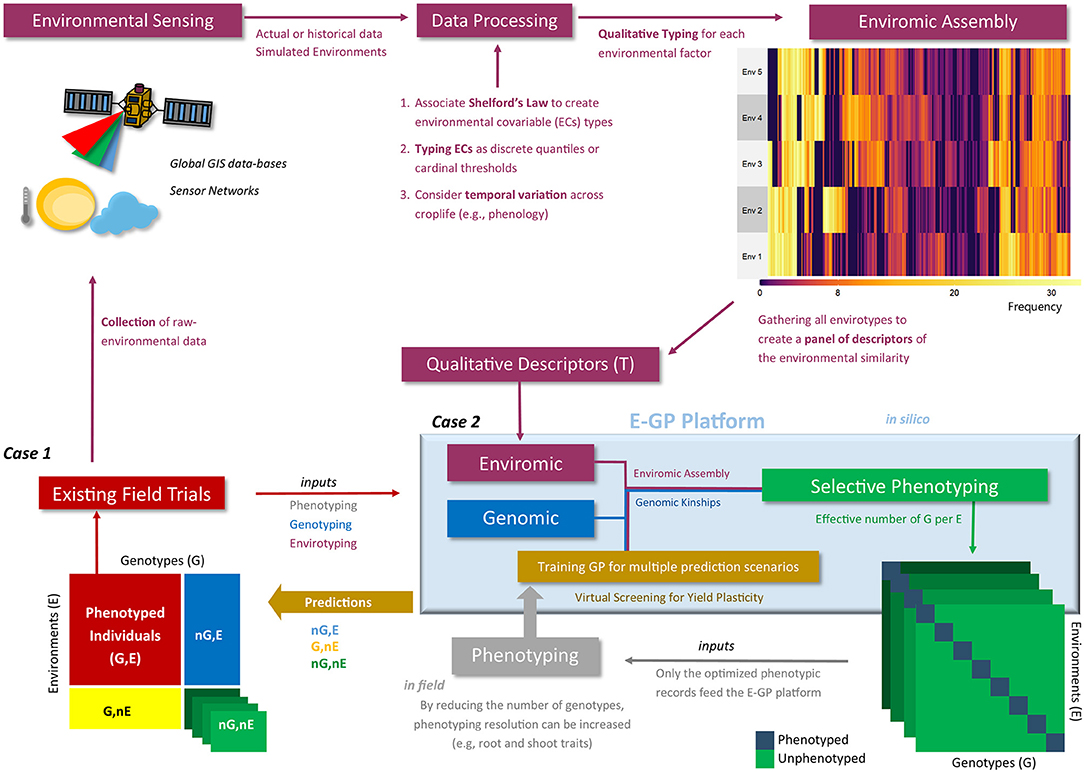
Figure | Workflow of the enviromic-aided genomic prediction (E-GP) considering the two study cases (Case 1 and Case 2) of this study.
Phenotypic records from existing field trials (red box) are based on observed genotypes (G) in tested growing environments (E). Currently, these data are being used for training prediction models considering untested genotypes at the same conditions (nG, E), especially when we have some type of structure of genetic relationships, such as genomic data (blue colors). In addition, novel growing conditions can be predicted (G, nE and nG, nE) using enviromic sources (wine colors), Case 1. First, raw environmental data are collected from trials involving equipment installed in situ (e.g., micro-weather stations) or remote sensing techniques. Then the raw data are processed and translated into an enviromic source that carries some ecophysiology process or statistical distribution of the raw data across time and space. The enviromic assembly is then finalized, in which its product is a matrix of envirotype markers by environments. Taking the T matrix as an example (qualitative descriptors based on typologies), a predictive breeding tool merging genomic, enviromics, and phenotypic data can be trained and deliver predictions for several scenarios of G × E. However, there is a second way to create an E-GP platform, the hereafter Case 2, in which the previously collected genomic and enviromic sources for a given TPE are used to develop in silico realizations of the expected G × E for a certain experimental network. Then, optimization algorithms are used to design a selective phenotyping strategy (green box) in which only the most representative genotype-environment combinations are phenotyped and considered for training the E-GP models (gray box). Finally, diverse G × E can also be predicted.





