October 09, 2025 | Crop Science |
Introduction: Traditional plant breeding often prioritizes explanatory models to understand biological mechanisms, which can limit the generalizability of selection decisions. Researchers from North Carolina State University (USA) and several Brazilian institutions address this gap by evaluating the shift toward prediction-based breeding. The study examines how tools like genomic selection (GS), high-throughput phenotyping (HTP), and enviromics can enhance genetic gain. Using a framework focused on operational utility, the authors review the role of stochastic simulations in optimizing breeding pipelines and reducing costs under dynamic agro-environmental scenarios.
Key findings: The review highlights a clear shift from explanation-oriented models toward prediction-driven breeding, arguing that for complex quantitative traits under climate variability, predictive reliability matters more than causal interpretation. Evidence synthesized across crops shows that prediction-based approaches—combining GE, stochastic simulation, HTP, and optimized trial design—can substantially increase genetic gain per unit time while reducing phenotyping costs. Simulation studies demonstrate that carefully designed training populations can achieve comparable prediction accuracy using only a small fraction of conventional field data, helping breeders anticipate overfitting risks before field deployment. The review further shows that integrating enviromics enables genotype recommendations across multiple environments by explicitly modeling genotype-by-environment (G×E) interactions through reaction norms. While advances in multi-omics and AI expand predictive power, the authors emphasize that data representativeness, environmental coverage, and institutional readiness—rather than model complexity alone—remain the main constraints to operationalizing prediction-based breeding for climate-resilient cultivar development.
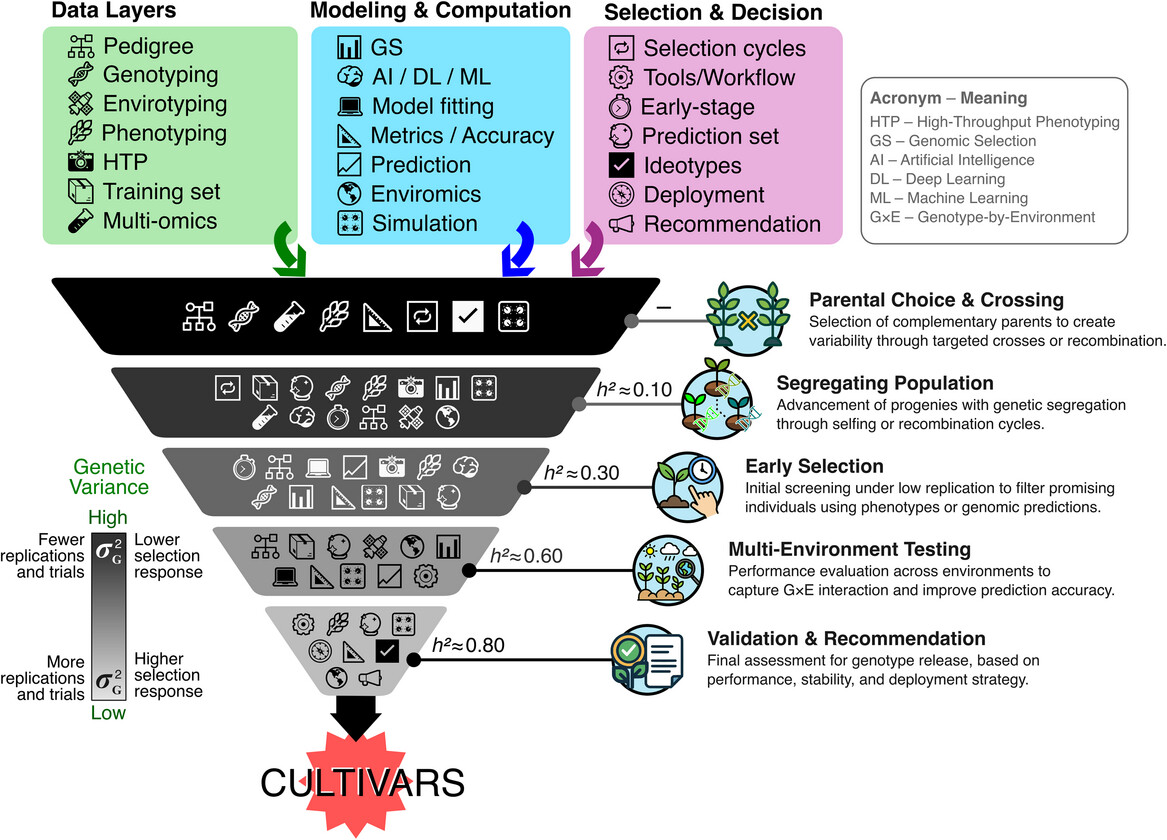
Figure | Funnel representation of the cultivar development pipeline and its main components (black and white flat icons inside each box). The five breeding stages are shown from top to bottom, illustrating the reduction in genetic variance (σG2 ) and the increase in heritability (h2) across the selection process. Each stage is linked to specific data layers (green), modeling tools (blue), and decision-making components (purple), all represented by flat icons. Icons are placed according to their most frequent or practical use, although most components could operate across multiple stages. The scale on the left highlights the decline in genetic variability and the increase in trial intensity. The pipeline converges in cultivar deployment, guided by data-driven decisions.





